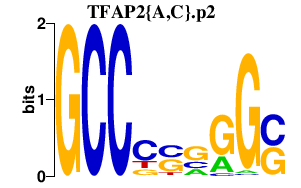
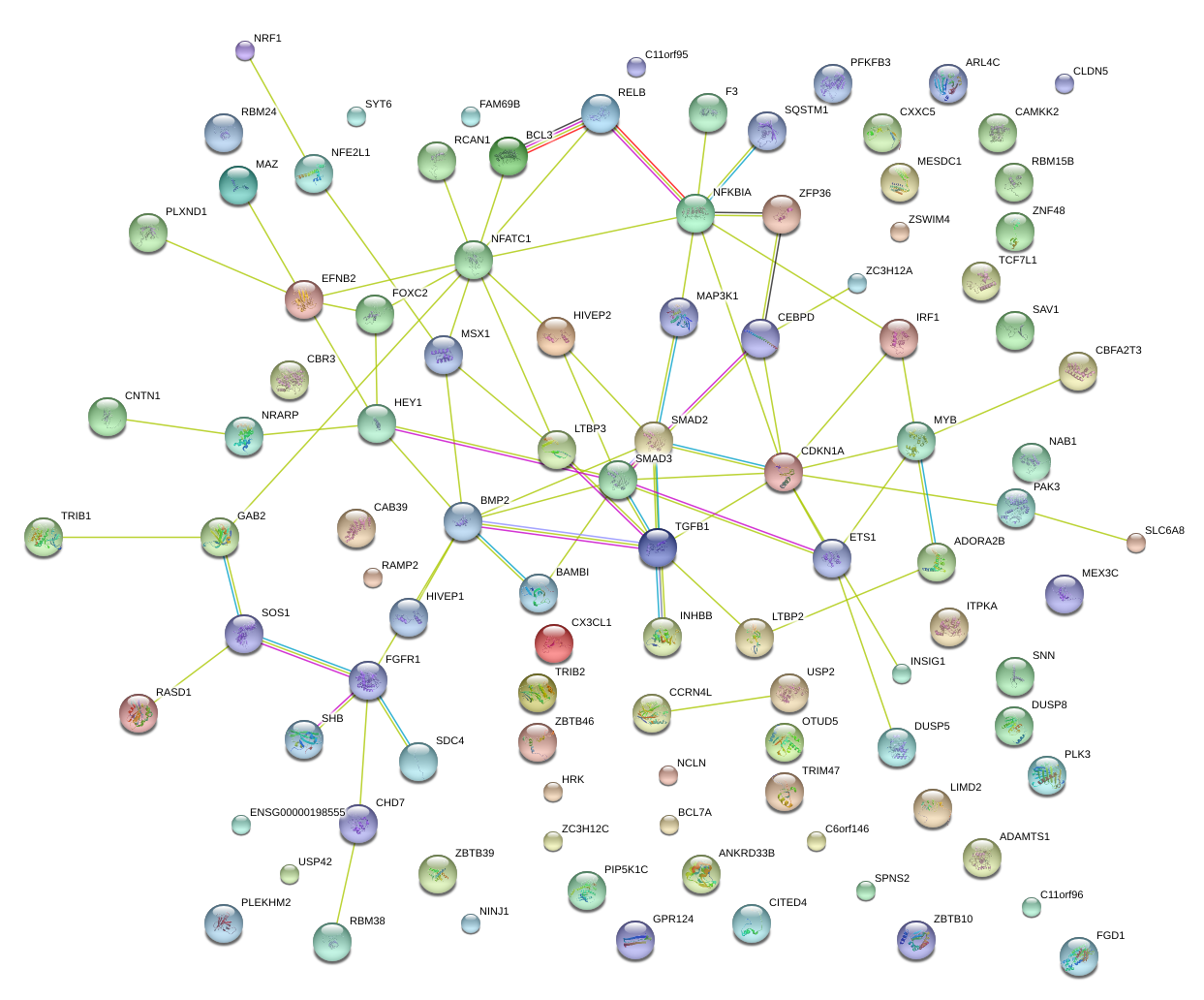
|
chr8_-_80679897
|
4.030
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr8_-_48650683
|
3.552
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_-_95007139
|
2.852
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr4_+_4861371
|
2.109
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr8_-_48650703
|
2.005
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_-_41328002
|
1.953
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr20_+_55966440
|
1.894
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr4_+_139936938
|
1.860
|
NM_012118
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr6_-_143266283
|
1.841
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr19_+_45251933
|
1.764
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr21_-_35987144
|
1.736
|
NM_004414
|
RCAN1
|
regulator of calcineurin 1
|
|
chr9_-_38069082
|
1.643
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr1_-_41328019
|
1.619
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr15_+_67358194
|
1.543
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr9_-_140196702
|
1.497
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr16_+_29817810
|
1.449
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr18_+_77155927
|
1.434
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr20_+_6748744
|
1.433
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr17_-_73874056
|
1.426
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr11_+_43963809
|
1.398
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr15_+_67358161
|
1.389
|
|
SMAD3
|
SMAD family member 3
|
|
chr14_-_35873823
|
1.347
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr20_-_43977023
|
1.272
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr17_-_73874634
|
1.257
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr15_+_41786055
|
1.255
|
NM_002220
|
ITPKA
|
inositol-trisphosphate 3-kinase A
|
|
chr15_+_81293286
|
1.228
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr12_-_117319231
|
1.216
|
NM_003806
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain)
|
|
chr6_-_4079390
|
1.195
|
NM_173563
|
C6orf146
|
chromosome 6 open reading frame 146
|
|
chr16_+_29817426
|
1.187
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr1_+_16010838
|
1.169
|
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr19_+_39897457
|
1.129
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr2_+_121103670
|
1.099
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr10_+_6186878
|
1.091
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr10_+_112257624
|
1.089
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr19_-_41859519
|
1.076
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr17_+_4402128
|
1.067
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr10_+_28966419
|
1.025
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr16_+_57406398
|
1.008
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr14_-_51134862
|
0.987
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr16_+_86600856
|
0.979
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr19_-_41859830
|
0.972
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr21_-_35986744
|
0.949
|
NM_203417
|
RCAN1
|
regulator of calcineurin 1
|
|
chr19_+_45504686
|
0.941
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chrX_-_54521866
|
0.917
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr18_+_77155714
|
0.901
|
NM_006162
NM_172388
NM_172390
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr19_-_41858948
|
0.896
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr2_+_85360373
|
0.895
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr10_+_6186758
|
0.887
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr11_-_1593149
|
0.884
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr10_+_6186903
|
0.871
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr12_+_122459791
|
0.866
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr7_+_129251579
|
0.860
|
|
NRF1
|
nuclear respiratory factor 1
|
|
chr1_+_16010816
|
0.858
|
NM_015164
|
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2
|
|
chr19_-_18391440
|
0.858
|
|
|
|
|
chr2_+_12856813
|
0.857
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chrX_-_48814758
|
0.856
|
|
OTUD5
|
OTU domain containing 5
|
|
chr19_-_41859253
|
0.852
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr5_-_131826426
|
0.841
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr18_+_77155966
|
0.838
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr19_-_41859332
|
0.832
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr11_-_128392061
|
0.823
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr19_+_3185927
|
0.817
|
|
NCLN
|
nicalin
|
|
chr12_-_121734543
|
0.812
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr21_+_37507209
|
0.809
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr2_-_208489695
|
0.792
|
NM_145280
|
METTL21A
|
methyltransferase like 21A
|
|
chr8_+_61591358
|
0.791
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr5_+_179247841
|
0.786
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr9_-_95896498
|
0.773
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr11_-_63536112
|
0.771
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr19_+_3185965
|
0.770
|
|
NCLN
|
nicalin
|
|
chr16_-_89007605
|
0.764
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr1_+_37940129
|
0.762
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr14_-_51135021
|
0.757
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr2_-_235405692
|
0.756
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr5_+_139027907
|
0.754
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr9_-_38069207
|
0.752
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr17_+_40913233
|
0.750
|
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr5_+_139028483
|
0.747
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr5_+_139027955
|
0.731
|
|
CXXC5
|
CXXC finger protein 5
|
|
chrX_-_54522318
|
0.722
|
NM_004463
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr1_+_45265896
|
0.720
|
|
PLK3
|
polo-like kinase 3
|
|
chr7_+_155089614
|
0.719
|
|
INSIG1
|
insulin induced gene 1
|
|
chr9_+_139606916
|
0.717
|
NM_152421
|
FAM69B
|
family with sequence similarity 69, member B
|
|
chr3_+_51428698
|
0.712
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr16_+_11762235
|
0.711
|
NM_003498
|
SNN
|
stannin
|
|
chr5_+_10564431
|
0.709
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr22_-_19511777
|
0.706
|
|
CLDN5
|
claudin 5
|
|
chr11_-_78128765
|
0.705
|
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr18_+_77155943
|
0.703
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr17_+_15848230
|
0.703
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chr17_-_17399494
|
0.695
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr19_+_13906207
|
0.693
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chrX_+_152953751
|
0.691
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr1_-_114696471
|
0.691
|
NM_001253772
|
SYT6
|
synaptotagmin VI
|
|
chr6_+_36646455
|
0.688
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr5_+_56111379
|
0.685
|
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr16_+_30406711
|
0.676
|
NM_152652
|
ZNF48
|
zinc finger protein 48
|
|
chr2_+_191513832
|
0.663
|
NM_005966
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1)
|
|
chr11_-_65325391
|
0.660
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr8_+_37654406
|
0.653
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr18_-_48723689
|
0.653
|
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr14_-_51135041
|
0.652
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr13_-_107187312
|
0.643
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr8_+_81398416
|
0.638
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr17_-_61777467
|
0.638
|
|
LIMD2
|
LIM domain containing 2
|
|
chr6_+_12012723
|
0.634
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr8_-_38325524
|
0.630
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr12_-_121734474
|
0.629
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr6_+_135502445
|
0.629
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr2_-_39348127
|
0.627
|
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr2_+_231577624
|
0.627
|
|
CAB39
|
calcium binding protein 39
|
|
chr19_-_3700466
|
0.624
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr8_+_126442557
|
0.621
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr11_+_109964086
|
0.618
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr19_+_3185860
|
0.613
|
NM_020170
|
NCLN
|
nicalin
|
|
chr7_+_6144589
|
0.610
|
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr5_+_139028025
|
0.605
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr12_-_57400199
|
0.605
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr22_-_19512001
|
0.604
|
|
CLDN5
|
claudin 5
|
|
chr11_-_119234628
|
0.603
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr20_-_62462568
|
0.599
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr21_-_28217272
|
0.596
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr3_-_129325175
|
0.590
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr4_+_1005609
|
0.590
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chrX_+_149531439
|
0.590
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr6_+_135502477
|
0.583
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr15_+_41221530
|
0.583
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr19_-_3700387
|
0.582
|
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr8_+_37654395
|
0.581
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr3_+_51428720
|
0.577
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr21_-_28217692
|
0.576
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr3_+_47866474
|
0.575
|
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr7_+_155250705
|
0.575
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr2_+_234263153
|
0.571
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr8_+_22423170
|
0.570
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr22_+_50354428
|
0.569
|
|
PIM3
|
pim-3 oncogene
|
|
chr19_+_10828772
|
0.569
|
|
DNM2
|
dynamin 2
|
|
chr2_+_231577536
|
0.566
|
NM_016289
|
CAB39
|
calcium binding protein 39
|
|
chr8_+_86089641
|
0.565
|
|
|
|
|
chr16_+_81478774
|
0.564
|
NM_198390
|
CMIP
|
c-Maf inducing protein
|
|
chr4_+_103422730
|
0.562
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr1_+_52608045
|
0.561
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr6_+_138188352
|
0.560
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_101767714
|
0.557
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr2_+_234263141
|
0.557
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr19_+_10828763
|
0.556
|
|
DNM2
|
dynamin 2
|
|
chr5_+_179248089
|
0.551
|
|
SQSTM1
|
sequestosome 1
|
|
chr11_-_118781612
|
0.549
|
NM_182557
|
BCL9L
|
B-cell CLL/lymphoma 9-like
|
|
chr15_-_101835360
|
0.548
|
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr18_-_45935627
|
0.546
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr1_+_1567556
|
0.546
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr16_-_57318452
|
0.545
|
|
PLLP
|
plasmolipin
|
|
chr1_-_224033383
|
0.544
|
|
TP53BP2
|
tumor protein p53 binding protein, 2
|
|
chr8_+_86089618
|
0.543
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr5_-_141257943
|
0.542
|
NM_002587
NM_032420
|
PCDH1
|
protocadherin 1
|
|
chr1_-_205290756
|
0.540
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr19_-_1863546
|
0.538
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr8_+_37654757
|
0.537
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr19_+_42788605
|
0.536
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr11_-_65326129
|
0.532
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr20_+_36531548
|
0.526
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr5_+_139027868
|
0.526
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr22_+_20067732
|
0.525
|
NM_001190326
NM_022720
|
DGCR8
|
DiGeorge syndrome critical region gene 8
|
|
chr1_-_33815498
|
0.525
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr19_+_10828813
|
0.523
|
|
DNM2
|
dynamin 2
|
|
chr7_-_139876718
|
0.522
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr2_+_171572698
|
0.522
|
|
SP5
|
Sp5 transcription factor
|
|
chr15_-_55880508
|
0.520
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr19_-_49865638
|
0.520
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr9_-_35732389
|
0.520
|
NM_006289
|
TLN1
|
talin 1
|
|
chr2_-_197457250
|
0.518
|
NM_020760
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2
|
|
chr1_-_33815411
|
0.517
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr9_+_82187599
|
0.516
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr16_+_67562704
|
0.513
|
NM_024519
NM_001193522
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr1_-_33430285
|
0.509
|
NM_001127361
NM_153341
|
RNF19B
|
ring finger protein 19B
|
|
chr19_+_10381763
|
0.509
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr11_+_64001974
|
0.509
|
NM_001243733
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr8_+_142138684
|
0.503
|
NM_014957
|
DENND3
|
DENN/MADD domain containing 3
|
|
chr20_+_1875825
|
0.501
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr14_+_104605059
|
0.501
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr8_-_10587942
|
0.500
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr9_+_116917716
|
0.499
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr1_+_100818052
|
0.499
|
|
CDC14A
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chr12_+_7037385
|
0.497
|
NM_001940
|
ATN1
|
atrophin 1
|
|
chr18_+_77160243
|
0.497
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_-_129035069
|
0.496
|
NM_006026
|
H1FX
|
H1 histone family, member X
|
|
chr6_+_36646489
|
0.496
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr11_+_369723
|
0.492
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr15_-_64338519
|
0.491
|
NM_014326
|
DAPK2
|
death-associated protein kinase 2
|
|
chr1_-_6545521
|
0.491
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr7_+_6144504
|
0.490
|
NM_032172
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr3_+_49027281
|
0.489
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr5_+_179247911
|
0.489
|
|
SQSTM1
|
sequestosome 1
|
|
chr10_-_3827418
|
0.483
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr19_+_49376488
|
0.480
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr17_+_40913211
|
0.478
|
NM_005854
|
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2
|
|
chr22_+_50354139
|
0.478
|
NM_001001852
|
PIM3
|
pim-3 oncogene
|
|
chr21_+_34775740
|
0.476
|
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1)
|
|
chr17_-_77813049
|
0.476
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr22_+_33197744
|
0.474
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|